tetrax.core package#
Subpackages#
Submodules#
tetrax.core.experimental_setup module#
- class tetrax.core.experimental_setup.CPWAntenna(width, gap, spacing)#
Bases:
tetrax.core.experimental_setup._AbstractMicrowaveAntennaCoplanar-waveguide antenna. The antenna is assumed to be infinitely thin.
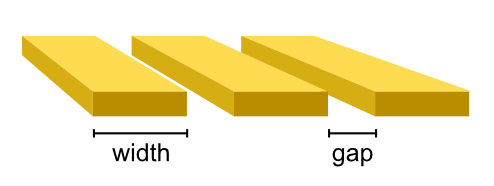
- Attributes
Methods
get_mesh
get_microwave_field_function
- get_mesh(span)#
- get_microwave_field_function(xyz)#
- class tetrax.core.experimental_setup.CurrentLoopAntenna(width, radius)#
Bases:
tetrax.core.experimental_setup._AbstractMicrowaveAntennaAntenna with the shape of a current loop.
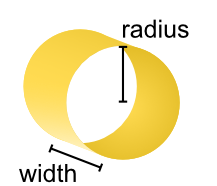
The current loop is oriented in the \(xy\) plane. For calculations, the current loop is taken to be infinitely thin in radial direction.
- Attributes
Methods
get_mesh
get_microwave_field_function
- get_mesh(span)#
- get_microwave_field_function(xyz)#
- class tetrax.core.experimental_setup.ExperimentalSetup(sample, name='my_experiment')#
Bases:
objectExperimental setup built around a
samplein which external parameters are defined and numerical experiments can be conducted.- Attributes
- sample
AbstractSample Sample object on which numerical experiments are conducted.
- name
str Name of the experimental setup, used to store results (default is
"my_experiment").- Bext
MeshVector Vector field of the static external magnetic field (given in T). Can also be set as a triplet, e.g. as
[0, 0, 0.5]for a homogeneous field of 500 mT in \(z\) direction.- antenna
_AbstractMicrowaveAntenna Microwave antenna in the experimental setup (see User Guide for details).
- sample
Methods
absorption([auto_eigenmodes, fmin, fmax, ...])Obtain the microwave absorption of the calculated eigensystem with respect to the microwave
antennain the experimental setup.eigenmodes([kmin, kmax, Nk, num_modes, k, ...])Calculate the spin-wave eigenmodes and their frequencies/dispersion of the sample in its current state using a dynamic matrix approach.
linewidths([auto_eigenmodes, k, verbose])Calculates the linewidths \(\Gamma=\alpha\epsilon\omega\) of spin-wave modes (with precession ellipticities \(\epsilon\)).
relax([tol, continue_with_least_squares, ...])Relax the magnetization
magof a sample in the experimental setup by minimizing the total magnetic energy.show([comp, scale, show_antenna, show_grid, ...])Shows the experimental setup, including external fields and microwave antennna.
show_mode([k, N, scale, on_equilibrium, ...])If spatial mode profiles are available, show a desired mode.
- property Bext#
- absorption(auto_eigenmodes=False, fmin=0, fmax=30, Nf=200, k=None, verbose=True)#
Obtain the microwave absorption of the calculated eigensystem with respect to the microwave
antennain the experimental setup.- Parameters
- auto_eigenmodesbool
If no eigenmodes are available in this experimental setup, calculate eigenmodes with default settings.
- fmin
float Minimum microwave frequency in GHz for which the absorption line is calculated (default is 0).
- fmax
float Maximum microwave frequency in GHz for which the absorption line is calculated (default is 30).
- Nf
int Number of microwave frequencies for which the absorption is calculated (default is 200).
- k
float If set to a value, the absorption line is calculated only for a single wave vector (default is
None). Also, the full dynamic susceptibility is saved to a dataframe. This argument is ignored in confined samples.- verbosebool
Output progress of the calculation (default is
True).
- Returns
- susceptibility
pandas.DataFrame Only if
kis notNoneor sample is a confined sample. Contains frequency-dependent real and imaginary part as well as magnitude as phase and magnitude of the averaged dynamic susceptiblity.- (absorbed_power, wave_vectors, frequencies)(
numpy.ndarray,numpy.ndarray,numpy.ndarray) If
kisNoneand sample is a waveguide or a layer sample. Saves the wave-vector and frequency- dependent microwave-power absorption (absorbed_power) together with the corresponding wave vectors and frequencies as a 2D arrays.
- susceptibility
Notes
The return of a wave-vector-dependent absorption calculation can be plotted using matplotlib as
>>> absorbed_power, wave_vectors, frequencies = exp.absorption(...) >>> import matplotlib.pyplot as plt >>> plt.pcolormesh(wave_vectors, frequencies, absorbed_power)
References
- 1
Verba, et al., “Damping of linear spin-wave modes in magnetic nanostructures: Local, nonlocal, and coordinate-dependent damping”, Phys. Rev. B 98, 104408 (2018)
- 2
Körber, et al., “Symmetry and curvature effects on spin waves in vortex-state hexagonal nanotubes”, Phys. Rev. B 104, 184429 (2021)
Examples
- property antenna#
- eigenmodes(kmin=- 40000000.0, kmax=40000000.0, Nk=81, num_modes=20, k=None, no_dip=False, h0_dip=False, num_cpus=1, save_modes=True, save_local=False, save_magphase=False, linewidths=False, perturbed_dispersion=False, save_stiffness_fields=False, verbose=True, save_disp=True, fname=None)#
Calculate the spin-wave eigenmodes and their frequencies/dispersion of the sample in its current state using a dynamic matrix approach.
The eigensystem is calculuated by numerically solving the linearized equation of motion of magnetizion (see User Guide for details). For waveguides or layer samples, a finite-element propagating-wave dynamic-matrix approach is used [1]. The dispersion is returned as a
pandas.DataFrame. Additionally, it is saved asdispersion.csv(ordispersion_perturbedmodes.csv, ifperturbed_dispersion=True) in the folder of the experimental setup.- Parameters
- kmin
float Minimum wave vector in rad/m (default is -40e6). This argument is ignored when calculating the eigensystem for confined samples.
- kmax
float Maximum wave vector in rad/m (default is 40e6). This argument is ignored when calculating the eigensystem for confined samples.
- Nk
int Number of wave-vector values, for which the mode frequencies are calculated. This argument is ignored when calculating the eigensystem for confined samples (default is 81).
- num_modes
int Determines how many modes (for confined samples) or branches of the dispersion (for waveguides and layers samples) are calculated in total (default is 20).
- k
floator array_like By supplying an
array_likeof wave vectors arbitrary k vector resolution is possible for particular or interesting parts of the dispersion. If set to a singlefloatvalue, the eigensystem is only calculated for a single wave vector (default isNone). This argument is ignored when calculating the eigensystem for confined samples.- no_dipbool
Exclude dipolar interaction from calculations (default is
False).- h0_dipbool
If
no_dipisTrue, still retain dipolar contributions in the equilibrium effective field.- num_cpus
int Number of CPU cores to be used in parallel for calculations (default is 1). If set to
-1, all available cores are used.- save_modesbool
Saves real and imaginary part of the mode profiles in cartesian basis as vtk files (default is
True). The mode profiles are saved in a/mode-profilessubdirectory in the folder of the experimental setup.- save_localbool
If
save_modesis alsoTrue, save the components of the mode profiles in the basis locally orthogonal to the equilibrum magnitization (default isFalse). The local components are saved in the samevtkfile as cartesian components.- save_magphasebool
If
save_modesis alsoTrue, save magnetide and phase of the mode-profile components (default isFalse). Saved in the same- linewidthsbool
Calculates the linewidths \(\Gamma=\alpha\epsilon\omega\) of the modes (with precession ellipticities \(\epsilon\)) and adds \(\Gamma/2\pi\) in GHz to the dataframe (default is
False).- perturbed_dispersionbool
After calculating the eigenmodes/normalmodes (possibly with
no_dip=True), calculate the zeroth-order pertubation of the dispersion including dynamic dipolar fields according to Ref [2]. Gives approximation for dispersion without dipolar hybridization (default isFalse). The results are appended to the returneddispersiondataframe.- save_stiffness_fieldsbool
If perturbed_dispersion is also
True, append the unitless individual stiffness fields (components) within the perturbed dispersion to thedispersiondataframe (default isFalse). Allows to numerically reverse engineer general spin-wave dispersions [2].- verbosebool
Output progress of the calculation (default is
True).- save_dispbool
Save the computed dispersion to a csv file in directory of the experimental setup (default is
True).- fname
str Filename for the dispersion file (default is
"dispersion.csv"ordispersion_perturbedmodes, depending onperturbed_dispersion).
- kmin
- Returns
- dispersion
pandas.DataFrame Dataframe (table) containing the calculated dispersion.
- dispersion
Notes
By default, the
dispersionDataFrame is structured ask (rad/m)
f0 (GHz)
f1 (GHz)
…
-40e6
1.426
2.352
…
…
…
…
…
In case
linewidths=True, the linewidths are added in units of GHz ask (rad/m)
f0 (GHz)
Gamma0/2pi (GHz)
…
-40e6
1.426
0.122
…
…
…
…
…
In case
perturbed_dispersion=True, additional columns are added ask (rad/m)
f0 (GHz)
f_pert_0 (GHz)
…
-40e6
1.426
484
…
…
…
…
…
Additionally, if
save_stiffnessfields=True, the unitless stiffness fields per magnetic interaction are also appendedk (rad/m)
f0 (GHz)
f_pert_0 (GHz)
Re(N21_exc)_0
…
-40e6
1.426
484
0.431
…
…
…
…
…
…
Indivual columns/rows can be obtained in the usual way for pandas.DataFrame`, for example, with
>>> k = dispersion["k (rad/m)"] >>> first_row = dispersion.iloc[0]
References
- 1
Körber, et al., “Finite-element dynamic-matrix approach for spin-wave dispersions in magnonic waveguides with arbitrary cross section”, AIP Advances 11, 095006 (2021)
- 2(1,2)
Körber and Kákay, “Numerical reverse engineering of general spin-wave dispersions: Bridge between numerics and analytics using a dynamic-matrix approach”, Phys. Rev. B 104, 174414 (2021)
Examples
- linewidths(auto_eigenmodes=False, k=None, verbose=True, **kwargs)#
Calculates the linewidths \(\Gamma=\alpha\epsilon\omega\) of spin-wave modes (with precession ellipticities \(\epsilon\)).
- Parameters
- auto_eigenmodesbool
If no modes have been previously calculated, calculates them automically (default is
False).- k
float Wave number at which the linewidths should be calculated. If None, the full wave-vector range available from the dispersion will be used (default is
None).- verbosebool
Output progress of the calculation (default is
True).- kwargs
Optional keyword arguments to be passed to the eigensolver (e.g.
num_cpus=-1).
- Returns
- linewidths
pandas.DataFrame Dataframe (table) containing the calculated linewidths (and the dispersion).
- linewidths
Notes
The line widths are returned together with the oscillation frequencies in units of GHz.
k (rad/m)
f0 (GHz)
Gamma0/2pi (GHz)
…
-40e6
1.426
0.122
…
…
…
…
…
References
- 1
Verba, et al., “Damping of linear spin-wave modes in magnetic nanostructures: Local, nonlocal, and coordinate-dependent damping”, Phys. Rev. B 98, 104408 (2018)
- 2
Körber, et al., “Symmetry and curvature effects on spin waves in vortex-state hexagonal nanotubes”, Phys. Rev. B 104, 184429 (2021)
Examples
- relax(tol=1e-12, continue_with_least_squares=False, return_last=False, save_mag=True, fname='mag.vtk', verbose=True)#
Relax the magnetization
magof a sample in the experimental setup by minimizing the total magnetic energy.- Parameters
- tol
float, optional Tolerance at which the minimization is considered successful (default is
1e-12).- continue_with_least_squaresbool
If minimization with conjugate-gradient method was not successful, continue with least-squares method (default is
False). This is more stable, but slower.- return_lastbool
If minimization was not successful, set the last iteration step as the sample magnetization (default is
False).- save_magbool
Save the resulting magnetization as vtk file to the folder of the experimental setup (default is
True).- fname
str Filename for magnetization file (default is
mag.vtk).- verbosebool
Output progress of the calculation (default is
True).
- tol
- Returns
- successbool
If the relaxation was successful of not.
Notes
For waveguide samples, the equilibration is not totally stable yet. Please use with care and check the resulting equilibrium states.
Examples
- show(comp='vec', scale=5, show_antenna=True, show_grid=False, show_extrusion=True)#
Shows the experimental setup, including external fields and microwave antennna.
- Parameters
- comp{“vec”, “x”, “y”, “z”}
If external field is nonzero, determines which component will be plotted.
- scale
float Determines the scale of the vector glyphs used to visualize the magnetization (default is 5).
- show_antennabool
If available, show the microwave antenna (default is
True).- show_gridbool
Show grid lines in plot (default is
False).- show_extrusionbool
Show full 3D mesh by extruding 1D or 2D mesh (default is
True).
- show_mode(k=0, N=0, scale=1, on_equilibrium=True, animated=False, periods=1, frames_per_period=20, fps=12, scale_mode=1)#
If spatial mode profiles are available, show a desired mode.
The mode is visualized as vector field which can either only show the dynamic component or the full magnetization (mode on-top of equilibrium). Setting
animated=Trueallows to animate the mode profiles by showing the oscillation over a desired number of periods (default is 1). The mode is automatically colored according to its magnitude.Warning
At the moment, animation is implemented such that it will play until the end and cannot be stopped (except by interrupting the kernel). So be careful with setting a large number of oscillation periods.
- Parameters
- k
float, optional Wave vector of desired mode in rad/µm (default is 0). This parameter will be ignored for confined samples.
- N
int, optional Mode/branch index of mode to be shown (default is 0).
- scale
float, optional Scaling of the mode (default is 1).
- on_equilibriumbool, optional
If true, show the mode on top of the equilibrium magnetization (default is True).
- animatedbool, optional
Make a mode movie by showing the magnetic oscillation over one period in a loop.
- periods
int, optional Number of oscillation periods to animate (default is 1). Will be ignored if
animated=False.- frames_per_period
int, optional Number of frames to animate per period (default is 20). Will be ignored if
animated=False.- fps
int, optional Frames per second for animation (default is 12).
- k
- class tetrax.core.experimental_setup.HomogeneousAntenna(theta=0, phi=0)#
Bases:
tetrax.core.experimental_setup._AbstractMicrowaveAntennaAntenna which produces a homogeneous microwave field.
- Attributes
Methods
get_mesh
get_microwave_field_function
- get_mesh()#
- get_microwave_field_function(xyz)#
- class tetrax.core.experimental_setup.StriplineAntenna(width, spacing)#
Bases:
tetrax.core.experimental_setup._AbstractMicrowaveAntennaStripline antenna. The antenna is assumed to be infinitely thin.
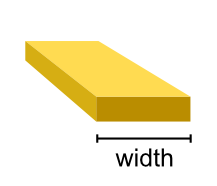
- Attributes
Methods
get_mesh
get_microwave_field_function
- get_mesh(span)#
- get_microwave_field_function(xyz)#
- tetrax.core.experimental_setup.create_experimental_setup(sample, name: str = 'my_experiment') tetrax.core.experimental_setup.ExperimentalSetup#
Creates an experimental setup around a given sample.
- Parameters
- sample
AbstractSample Sample object to create ExperimentalSetup around.
- name
str Name of the experimental setup.
- sample
- Returns
tetrax.core.mesh module#
This core submodule provides several subclasses of np.ndarray to define different scalar and vector fields on meshes. The import convention
>>> import numpy as np
is used.
- class tetrax.core.mesh.FlattenedAFMMeshVector(input_array)#
Bases:
numpy.ndarrayClass of shape for antiferromagnets (6*nx,) to hold the flattened vector fields of each sublattice defined on the same mesh.
The entries of the vector are ordered according to
\[\mathbf{a} = (a_{x_1}, ... , a_{x_N}, a_{y_1}, ... , a_{y_N}, a_{z_1}, ... , a_{z_N}, b_{x_1}, ... , b_{x_N}, b_{y_1}, ... , b_{y_N}, b_{z_1}, ... , b_{z_N})\]while \(N\) = nx and \(\mathbf{a}\), \(\mathbf{b}\) are the vector fields of the respective sublattices. The input array_like needs to be one dimensional. The number of nodes nx is inferred from the input array.
- Attributes
- nx
int Number of nodes.
- x1
MeshScalar The \(x\) component of the first sublattice vector field at each mesh node.
- y1
MeshScalar The \(y\) component of the first sublattice vector field at each mesh node.
- z1
MeshScalar The \(z\) component of the first sublattice vector field at each mesh node.
- x2
MeshScalar The \(x\) component of the second sublattice vector field at each mesh node.
- y2
MeshScalar The \(y\) component of the second sublattice vector field at each mesh node.
- z2
MeshScalar The \(z\) component of the second sublattice vector field at each mesh node.
- nx
Methods
to_two_unflattened()
Returns the indivudal vector fields of the sublattices as two separate :py:class:`MeshVector`s.
to_spherical()
Converts the vectors on each sublattice to spherical coordinates (only angles), returning a
FlattenedAFMMeshVectorSpherical.- to_spherical()#
- to_two_unflattened()#
- property x1#
- property x2#
- property y1#
- property y2#
- property z1#
- property z2#
- class tetrax.core.mesh.FlattenedAFMMeshVectorSpherical(input_array)#
Bases:
numpy.ndarrayClass for antiferromagnets of shape (4*nx,) holding two two-component unit-vector fields of shape (2*nx,) each defined on a mesh using spherical angles.
The entries of the vector field are ordered according to
\[\mathbf{v} = (\theta_{1,1}, ... , \theta_{1,N}, \varphi_{1,1}, ... , \varphi_{1,N},\theta_{2,1}, ... , \theta_{2,N}, \varphi_{2,1}, ... , \varphi_{2,N})\]while \(N\) = nx and \(\theta_{i,j}\) and \(\varphi_{i,j}\) being polar and azimuthal angle of the unit-vector field of the \(i\) th sublattice at the \(j\) th node. The input array_like needs to be one dimensional. The number of nodes nx is inferred from shape[0] of the input array.
See also
- Attributes
- nx
int Number of nodes.
- theta1
MeshScalar The polar angle (\(\theta_1\)) of the unit-vector field of the first sublattice at each mesh node.
- phi1
MeshScalar The azimuthal angle (\(\varphi_1\)) of the unit-vector of the first sublattice field at each mesh node.
- theta2
MeshScalar The polar angle (\(\theta_2\)) of the unit-vector field of the first sublattice at each mesh node.
- phi2
MeshScalar The azimuthal angle (\(\varphi_2\)) of the unit-vector of the first sublattice field at each mesh node.
- nx
Methods
to_cartesian()
Returns the vector fields for both sublattices as a flattened
FlattenedAFMMeshVectorin cartesian coordinates.- property phi1#
- property phi2#
- property theta1#
- property theta2#
- to_cartesian()#
- class tetrax.core.mesh.FlattenedLocalAFMMeshVector(input_array)#
Bases:
numpy.ndarrayClass for antiferromagnets of shape (4*nx,) to hold the flattened vector fields of each sublattice defined on the same mesh.
This class is used to describe vector fields locally ortoghonal and expressed in the \((u,v,w)\) basis attached to the equilibrium magnetization of each sublattice, hence the name. Because of that, the third (\(w\)) compontent of the vector field of each sublattice is always zero and therefore omitted. The entries of the vector are ordered according to
\[\mathbf{v} = (u_{1,1}, ... , u_{1,N}, v_{1,1}, ... , v_{1,N}, u_{2,1}, ... , u_{2,N}, v_{2,1}, ... , v_{2,N})\]- while \(N\) = nx and \(u_{i,j}\) and \(v_{i,j}\) being locally orthogonal components
of the \(i\) th sublattice at the \(j\) th node.
The input array_like needs to be one dimensional. The number of nodes nx is inferred from the input array.
- Attributes
- nx
int Number of nodes.
- u1
MeshScalar The \(u\) component of the first sublattice vector field at each mesh node.
- v1
MeshScalar The \(v\) component of the first sublattice vector field at each mesh node.
- u1
MeshScalar The \(u\) component of the first sublattice vector field at each mesh node.
- v2
MeshScalar The \(v\) component of the second sublattice vector field at each mesh node.
- nx
Methods
all([axis, out, keepdims, where])Returns True if all elements evaluate to True.
any([axis, out, keepdims, where])Returns True if any of the elements of a evaluate to True.
argmax([axis, out, keepdims])Return indices of the maximum values along the given axis.
argmin([axis, out, keepdims])Return indices of the minimum values along the given axis.
argpartition(kth[, axis, kind, order])Returns the indices that would partition this array.
argsort([axis, kind, order])Returns the indices that would sort this array.
astype(dtype[, order, casting, subok, copy])Copy of the array, cast to a specified type.
byteswap([inplace])Swap the bytes of the array elements
choose(choices[, out, mode])Use an index array to construct a new array from a set of choices.
clip([min, max, out])Return an array whose values are limited to
[min, max].compress(condition[, axis, out])Return selected slices of this array along given axis.
conj()Complex-conjugate all elements.
conjugate()Return the complex conjugate, element-wise.
copy([order])Return a copy of the array.
cumprod([axis, dtype, out])Return the cumulative product of the elements along the given axis.
cumsum([axis, dtype, out])Return the cumulative sum of the elements along the given axis.
diagonal([offset, axis1, axis2])Return specified diagonals.
dump(file)Dump a pickle of the array to the specified file.
dumps()Returns the pickle of the array as a string.
fill(value)Fill the array with a scalar value.
flatten([order])Return a copy of the array collapsed into one dimension.
getfield(dtype[, offset])Returns a field of the given array as a certain type.
item(*args)Copy an element of an array to a standard Python scalar and return it.
itemset(*args)Insert scalar into an array (scalar is cast to array's dtype, if possible)
max([axis, out, keepdims, initial, where])Return the maximum along a given axis.
mean([axis, dtype, out, keepdims, where])Returns the average of the array elements along given axis.
min([axis, out, keepdims, initial, where])Return the minimum along a given axis.
newbyteorder([new_order])Return the array with the same data viewed with a different byte order.
nonzero()Return the indices of the elements that are non-zero.
partition(kth[, axis, kind, order])Rearranges the elements in the array in such a way that the value of the element in kth position is in the position it would be in a sorted array.
prod([axis, dtype, out, keepdims, initial, ...])Return the product of the array elements over the given axis
ptp([axis, out, keepdims])Peak to peak (maximum - minimum) value along a given axis.
put(indices, values[, mode])Set
a.flat[n] = values[n]for all n in indices.ravel([order])Return a flattened array.
repeat(repeats[, axis])Repeat elements of an array.
reshape(shape[, order])Returns an array containing the same data with a new shape.
resize(new_shape[, refcheck])Change shape and size of array in-place.
round([decimals, out])Return a with each element rounded to the given number of decimals.
searchsorted(v[, side, sorter])Find indices where elements of v should be inserted in a to maintain order.
setfield(val, dtype[, offset])Put a value into a specified place in a field defined by a data-type.
setflags([write, align, uic])Set array flags WRITEABLE, ALIGNED, WRITEBACKIFCOPY, respectively.
sort([axis, kind, order])Sort an array in-place.
squeeze([axis])Remove axes of length one from a.
std([axis, dtype, out, ddof, keepdims, where])Returns the standard deviation of the array elements along given axis.
sum([axis, dtype, out, keepdims, initial, where])Return the sum of the array elements over the given axis.
swapaxes(axis1, axis2)Return a view of the array with axis1 and axis2 interchanged.
take(indices[, axis, out, mode])Return an array formed from the elements of a at the given indices.
tobytes([order])Construct Python bytes containing the raw data bytes in the array.
tofile(fid[, sep, format])Write array to a file as text or binary (default).
tolist()Return the array as an
a.ndim-levels deep nested list of Python scalars.tostring([order])A compatibility alias for tobytes, with exactly the same behavior.
trace([offset, axis1, axis2, dtype, out])Return the sum along diagonals of the array.
transpose(*axes)Returns a view of the array with axes transposed.
var([axis, dtype, out, ddof, keepdims, where])Returns the variance of the array elements, along given axis.
view([dtype][, type])New view of array with the same data.
dot
- property u1#
- property u2#
- property v1#
- property v2#
- class tetrax.core.mesh.FlattenedLocalMeshVector(input_array)#
Bases:
numpy.ndarrayClass for two-component vector fields of shape (2*nx,) defined on a mesh.
This class is used to describe vector fields locally ortoghonal and expressed in the \((u,v,w)\) basis attached to the equilibrium magnetization, hence the name. Because of that, the third (\(w\)) compontent of the vector field is always zero and therefore omitted. The entries of the vector field are ordered according to
\[\mathbf{a} = (a_{u_1}, ... , a_{u_N}, a_{v_1}, ... , a_{v_N})\]while \(N\) = nx. The input array_like needs to be one dimensional. The number of nodes nx is inferred from shape[0] of the input array.
- Attributes
- nx
int Number of nodes.
- u
MeshScalar The first (\(u\)) component of the vector field at each mesh node.
- v
MeshScalar The second (\(v\)) component of the vector field at each mesh node.
- nx
Methods
to_unflattened()
Returns the vector field as an unflattened
LocalMeshVector.- to_unflattened()#
- property u#
- property v#
- class tetrax.core.mesh.FlattenedMeshVector(input_array)#
Bases:
numpy.ndarrayClass for three-component vector fields of shape (3*nx,) defined on a mesh.
The entries of the vector are ordered according to
\[\mathbf{a} = (a_{x_1}, ... , a_{x_N}, a_{y_1}, ... , a_{y_N}, a_{z_1}, ... , a_{z_N})\]while \(N\) = nx. The input array_like needs to be one dimensional and cannot be a
FlattenedLocalMeshVector. The number of nodes nx is inferred from the length of the input array.- Attributes
- nx
int Number of nodes.
- x
MeshScalar The \(x\) component of the vector field at each mesh node.
- y
MeshScalar The \(y\) component of the vector field at each mesh node.
- z
MeshScalar The \(z\) component of the vector field at each mesh node.
- nx
Methods
to_ùnflattened()
Returns the vector field as an unflattened
MeshVector.to_spherical()
Converts the vector to spherical coordinates (only angles), returning a
FlattenedMeshVectorSpherical.- to_spherical()#
- to_unflattened()#
- property x#
- property y#
- property z#
- class tetrax.core.mesh.FlattenedMeshVectorSpherical(input_array)#
Bases:
numpy.ndarrayClass for two-component unit-vector fields of shape (2*nx,) defined on a mesh using spherical angles.
The entries of the vector field are ordered according to
\[\mathbf{v} = (\theta_1, ... , \theta_N, \varphi_1, ... , \varphi_N)\]while \(N\) = nx and \(\theta_i\) and \(\varphi_i\) being polar and azimuthal angle of the unit-vector field. The input array_like needs to be one dimensional. The number of nodes nx is inferred from shape[0] of the input array.
See also
- Attributes
- nx
int Number of nodes.
- theta
MeshScalar The polar angle (\(\theta\)) of the unit-vector field at each mesh node.
- phi
MeshScalar The azimuthal angle (\(\varphi\)) of the unit-vector field at each mesh node.
- nx
Methods
to_cartesian()
Returns the vector field as a flattened
FlattenedMeshVectorin cartesian coordinates.- property phi#
- property theta#
- to_cartesian()#
- class tetrax.core.mesh.LocalMeshVector(input_array)#
Bases:
numpy.ndarrayClass for two-component vector fields of shape (nx, 2) defined on a mesh.
This class is used to describe vector fields locally ortoghonal and expressed in the \((u,v,w)\) basis attached to the equilibrium magnetization, hence the name. Because of that, the third (\(w\)) compontent of the vector field is always zero and therefore omitted. The entries of the vector field are ordered according to
\[\begin{split}\mathbf{a} = \begin{pmatrix} a_{u_1} & a_{v_1} \\ & \vdots & \\ a_{u_N} & a_{v_N} \\ \end{pmatrix}\end{split}\]while \(N\) = nx. The input array_like needs to be one dimensional and cannot be a
FlattenedMeshVector. The number of nodes nx is inferred from the length of the input array.See also
- Attributes
- nx
int Number of nodes.
- u
MeshScalar The first (\(u\)) component of the vector field at each mesh node.
- v
MeshScalar The second (\(v\)) component of the vector field at each mesh node.
- nx
Methods
to_flattened()
Returns the vector field as a
FlattenedLocalMeshVector.- to_flattened()#
- property u#
- property v#
- class tetrax.core.mesh.MeshScalar(input_array)#
Bases:
numpy.ndarrayClass for scalar fields defined on a mesh.
Input array_like needs to be one dimensional. The number of nodes nx is inferred from the length of the input array.
- Attributes
- nx
int Number of nodes.
- nx
Methods
all([axis, out, keepdims, where])Returns True if all elements evaluate to True.
any([axis, out, keepdims, where])Returns True if any of the elements of a evaluate to True.
argmax([axis, out, keepdims])Return indices of the maximum values along the given axis.
argmin([axis, out, keepdims])Return indices of the minimum values along the given axis.
argpartition(kth[, axis, kind, order])Returns the indices that would partition this array.
argsort([axis, kind, order])Returns the indices that would sort this array.
astype(dtype[, order, casting, subok, copy])Copy of the array, cast to a specified type.
byteswap([inplace])Swap the bytes of the array elements
choose(choices[, out, mode])Use an index array to construct a new array from a set of choices.
clip([min, max, out])Return an array whose values are limited to
[min, max].compress(condition[, axis, out])Return selected slices of this array along given axis.
conj()Complex-conjugate all elements.
conjugate()Return the complex conjugate, element-wise.
copy([order])Return a copy of the array.
cumprod([axis, dtype, out])Return the cumulative product of the elements along the given axis.
cumsum([axis, dtype, out])Return the cumulative sum of the elements along the given axis.
diagonal([offset, axis1, axis2])Return specified diagonals.
dump(file)Dump a pickle of the array to the specified file.
dumps()Returns the pickle of the array as a string.
fill(value)Fill the array with a scalar value.
flatten([order])Return a copy of the array collapsed into one dimension.
getfield(dtype[, offset])Returns a field of the given array as a certain type.
item(*args)Copy an element of an array to a standard Python scalar and return it.
itemset(*args)Insert scalar into an array (scalar is cast to array's dtype, if possible)
max([axis, out, keepdims, initial, where])Return the maximum along a given axis.
mean([axis, dtype, out, keepdims, where])Returns the average of the array elements along given axis.
min([axis, out, keepdims, initial, where])Return the minimum along a given axis.
newbyteorder([new_order])Return the array with the same data viewed with a different byte order.
nonzero()Return the indices of the elements that are non-zero.
partition(kth[, axis, kind, order])Rearranges the elements in the array in such a way that the value of the element in kth position is in the position it would be in a sorted array.
prod([axis, dtype, out, keepdims, initial, ...])Return the product of the array elements over the given axis
ptp([axis, out, keepdims])Peak to peak (maximum - minimum) value along a given axis.
put(indices, values[, mode])Set
a.flat[n] = values[n]for all n in indices.ravel([order])Return a flattened array.
repeat(repeats[, axis])Repeat elements of an array.
reshape(shape[, order])Returns an array containing the same data with a new shape.
resize(new_shape[, refcheck])Change shape and size of array in-place.
round([decimals, out])Return a with each element rounded to the given number of decimals.
searchsorted(v[, side, sorter])Find indices where elements of v should be inserted in a to maintain order.
setfield(val, dtype[, offset])Put a value into a specified place in a field defined by a data-type.
setflags([write, align, uic])Set array flags WRITEABLE, ALIGNED, WRITEBACKIFCOPY, respectively.
sort([axis, kind, order])Sort an array in-place.
squeeze([axis])Remove axes of length one from a.
std([axis, dtype, out, ddof, keepdims, where])Returns the standard deviation of the array elements along given axis.
sum([axis, dtype, out, keepdims, initial, where])Return the sum of the array elements over the given axis.
swapaxes(axis1, axis2)Return a view of the array with axis1 and axis2 interchanged.
take(indices[, axis, out, mode])Return an array formed from the elements of a at the given indices.
tobytes([order])Construct Python bytes containing the raw data bytes in the array.
tofile(fid[, sep, format])Write array to a file as text or binary (default).
tolist()Return the array as an
a.ndim-levels deep nested list of Python scalars.tostring([order])A compatibility alias for tobytes, with exactly the same behavior.
trace([offset, axis1, axis2, dtype, out])Return the sum along diagonals of the array.
transpose(*axes)Returns a view of the array with axes transposed.
var([axis, dtype, out, ddof, keepdims, where])Returns the variance of the array elements, along given axis.
view([dtype][, type])New view of array with the same data.
dot
- class tetrax.core.mesh.MeshVector(input_array)#
Bases:
numpy.ndarrayClass for three-component vector fields of shape (nx, 3) defined on a mesh.
The entries of the vector are ordered according to
\[\begin{split}\mathbf{a} = \begin{pmatrix} a_{x_1} & a_{y_1} & a_{z_1} \\ & \vdots & \\ a_{x_N} & a_{y_N} & a_{z_N} \\ \end{pmatrix}\end{split}\]while \(N\) = nx. The input array_like needs to be two dimensional with shape[1] = 3 (array of triplets). The number of nodes nx is inferred from shape[0] of the input array.
- Attributes
- nx
int Number of nodes.
- x
MeshScalar The \(x\) component of the vector field at each mesh node.
- y
MeshScalar The \(y\) component of the vector field at each mesh node.
- z
MeshScalar The \(z\) component of the vector field at each mesh node.
- nx
Methods
to_flattened()
Returns the vector field as
FlattenedMeshVector.- to_flattened()#
- property x#
- property y#
- property z#
- tetrax.core.mesh.flattened_spherical_to_cartesian_afm(vec, nx)#
Convert a flattened mesh vector from spherical angles to cartesian coordinates.
This method exists solely for cases where mesh-vector data types are overwritten by
numpy.asarray(). This is the case for example inscipy.optimize.minimize().- Parameters
- vecarray_like
Array assumed to be of length 4*nx (for antiferromagnets) containing the spherical angles of both sublattices Checks are not done to save time.
- nx
int Number of nodes.
- Returns
numpy.arrayArray of length 6*nx containing the cartesian coordinates.
See also
- tetrax.core.mesh.flattened_spherical_to_cartesian_fm(vec, nx)#
Convert a flattened mesh vector from spherical angles to cartesian coordinates.
This method exists solely for cases where mesh-vector data types are overwritten by
numpy.asarray(). This is the case for example inscipy.optimize.minimize().- Parameters
- vecarray_like
Array assumed to be of length 2*nx (for ferromagnets) containing the spherical angles. Checks are not done to save time.
- nx
int Number of nodes.
- Returns
numpy.arrayArray of length 3*nx containing the cartesian coordinates.
See also
- tetrax.core.mesh.make_flattened_AFM(a, b)#
Creates a
FlattenedAFMMeshVectorfrom two individual :py:class:`MeshVector`s.- Parameters
- a
MeshVector Vector field of the first sublattice.
- b
MeshVector Vector field of the second sublattice.
- a
- Returns
FlattenedAFMMeshVectorFlattened vector field of the AFM lattice.
See also
tetrax.core.operators module#
tetrax.core.sample module#
- class tetrax.core.sample.AbstractSample(name, geometry, magnetic_order, dim)#
Bases:
abc.ABCBase class for the concrete sample classes
_ConfinedSampleFM_ConfinedSampleAFM_WaveguideSampleFM_WaveguideSampleAFM_LayerSampleFM_LayerSampleAFM
which are created by multiple-inheritance from this base class together with mixin classes for magnetic order (FM or AFM) and geometry type (confined, waveguide, layer). The available attributes for material parameters depend on the magnetic order (see User Guide for details).
- Attributes
- name
str Name of the sample.
- nx
int Total number of nodes in the mesh of the sample.
- nb
int Number of boundary nodes in the mesh of the sample.
- mesh
meshio.Mesh Mesh of the sample.
- xyz
MeshVector Coordinates on the mesh of the sample.
- scale
float Scale of the sample (default is 1e-9, such that all spatial dimensions are given in nanometer).
- name
Methods
average(field)Calculate the average of a scalar of vector field within the given sample.
field_to_file(field[, fname, qname])Calls the
tetrax.helpers.io.write_field_to_file()function to write scalar- or vector field to a file.plot(fields[, comp, scale, labels])Plot a vector or scalar field which is defined on each node of the mesh.
read_mesh(fname)Read a mesh from a file using meshio and set it as the geometry of a sample.
scan_along_curve(field, curve[, num_points, ...])Interpolate a given scalar or vector field along a specified curve.
set_geom(mesh)Set the geometry/mesh of the sample and do neccesary preprocessing for later calculations.
show([show_node_labels, show_mag, comp, ...])Show the sample.
- average(field)#
Calculate the average of a scalar of vector field within the given sample.
- field_to_file(field, fname='field.vtk', qname=None)#
Calls the
tetrax.helpers.io.write_field_to_file()function to write scalar- or vector field to a file.
- plot(fields, comp='vec', scale=5, labels=None)#
Plot a vector or scalar field which is defined on each node of the mesh. Multiple fields can be plotted at the same time.
- Parameters
- fields
numpy.arrayorlist(numpy.array) Scalar field of shape (nx,), vector field of shape (nx,3) or list containing any choice of the aforementioned.
- comp{“vec”, “x”, “y”, “z”}
If field is a vector field, this parameter determines which component will be plotted.
- scale
float Determines the scale of the vector glyphs used to visualize a vectorfield (default is 5).
- labels
strorlist(str) Label(s) of the vectorfield(s) to be plotted. If
labelsis a list, it needs to have the same length asfields(default isNone).
- fields
- read_mesh(fname)#
Read a mesh from a file using meshio and set it as the geometry of a sample.
- Parameters
- fname
str Name of the mesh file.
- fname
- scan_along_curve(field, curve, num_points=100, return_curve=False)#
Interpolate a given scalar or vector field along a specified curve.
- Parameters
- field
MeshScalarorMeshVector Field to be interpolated.
- curve
ndarray,shape(nx, 3) or tuple/list ofshape(2, 3) The points along the interpolation curve or the start and end points of the curve
((x1, y1, z1),(x2, y2, z2)).- num_points
int The number of points to interpolate along the curve. Will only be considered if start and end points are given (default is 100).
- return_curvebool
Next to the interpolated values, also return the interpolation curve.
- field
- Returns
- interp_values
ndarray,shape(num_points,m) The interpolated values along the curve with
mbeing the size of the second dimension offield.
- interp_values
Examples
- set_geom(mesh)#
Set the geometry/mesh of the sample and do neccesary preprocessing for later calculations.
According to the dimension of the sample, the mesh is checked for compatibility. After sucessefully setting the mesh and extrancting necessary information, such as the number of nodes
nx, the coordinates of the mesh verticesxyzor the normal vectorsnv, the discretized differential operators (poisson,div_x, …) are calculated and the magnetic tensors are set up.- Parameters
- mesh
meshio.Mesh
- mesh
- abstract show(show_node_labels=False, show_mag=True, comp='vec', scale=5, show_scaled_mag=True, show_grid=False, show_extrusion=True)#
Show the sample. Displays the mesh and, if available, the magnetization(s).
- Parameters
- show_node_labelsbool
If true, shows the label associated to each node.
- show_magbool
Show the magnetization (or magnetizations, in the case of antiferromagnetic samples).
- comp{“vec”, “x”, “y”, “z”}
Determines which component will be plotted.
- scale
float Determines the scale of the vector glyphs used to visualize the magnetization (default is 5).
- show_scaled_magbool
Scale magnetization with spatially dependent saturation magnetization (default is
True).- show_gridbool
Show background grid (default is
False).- show_extrusionbool
Show full 3D mesh by extruding 1D or 2D mesh (default is
True).
- Returns
- plot
k3d_plot_object
- plot
- tetrax.core.sample.create_sample(name='my_sample', geometry='waveguide', magnetic_order='FM')#
Create a sample with certain geometry and magnetic order.
- Parameters
- Returns
- sample
AbstractSample
- sample